CRISPR Gene Editing
Genomic Editing with CRISPR/CAS9
- CAS9 expression lentivirus;
- Integration Defective Cas9 Lentivirus;
- CAS9 Adenovirus;
- gRNA lentivector cloning kits;
- Epigenomic: CRISPRi and CRISPRa;
Targeted and precise genomic gene-editing technologies are the tools for genomic correction, modification, and gene therapy. The TALEN, ZFN, and CRISPR/Cas are the three main genome editing technologies.
The lately discovered gene-editing technology, the CRISPR/Cas (Clustered Regularly Interspaced Short Palindromic Repeats) technology has (1) higher targeting accuracy; (2) much more target sequence selection; (3) much less complexity; and (4) much less off-target cell toxicity than the previous genome editing technologies: TALEN (transcription activator-like effector nuclease) and ZEN (Zinc-finger nuclease). The CRISPR/Cas and CRISPR-associated (Cas) endonucleases were discovered in E Coli and have since been developed into a powerful technology for genomic editing, making possible highly specific genome modifications far more quickly and economically.
CRISPR/Cas gene editing requires three components:
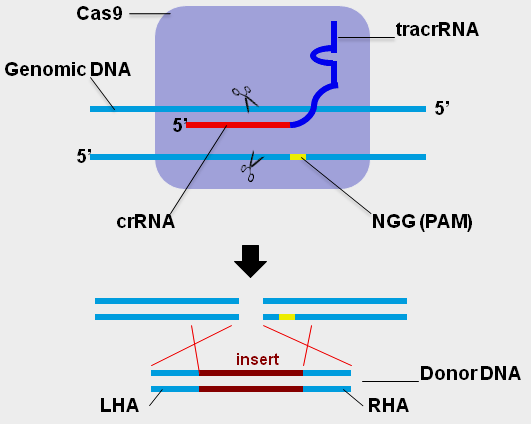
1) Target specific guild RNA (gRNA): it comprises two segments: a targeting sequence (crRNA) containing the target complementary RNA, and an auxiliary trans-activating non-coding RNA sequence (tracrRNA).
2) Cas9 endonuclease: The gRNA binds to Cas9 enzyme to the formation of a gRNA-Cas9 complex that will bind to target specific genomic loci and cleave the corresponding genomic DNA target sequence.
3) The Donor DNA template (“knock In”): For genomic modification application, a double-strand or single-stranded repair DNA is required after the Cas9 creates the double-stranded breaks at desired genomic loci. (Note: when this Donor DNA is not present, the CRISPR only creates the knockout).
CRISPR target site selection (for gRNA sequence):
The selection of the target sequence within the gene of interest is critical to the efficacy and specificity of genetic editing with CRISPR/Cas9. The crRNA segment of the gRNA will only bind to DNA targets that are immediately upstream of the proper Protospacer Adjacent Motif (PAM) sequence, which for CRISPR/Cas9 is NGG. The target sequence (20bp ~ 30bp) can be in either the sense or anti-sense orientation with respect to the target gene. It is a good idea to create several target sequences for your gene of interest and to select sequences with minimal homology to other genes, in order to find a sequence with good cleavage efficiency and minimal off-target effects. (See the links at the bottom of the page to online bioinformatics tools to assist in selecting a gRNA sequence with minimal off-target effects.
Online tools for target sequence selection: 5′- “(20-30 target sequence) + PAM (NGG)”
(Note: the selected sequences are in front of the NGG in genomic sequence, but NGG should not be included in the synthesized gRNA)
http://zifit.partners.org/ZiFiT/Introduction.aspx
http://www.e-crisp.org/E-CRISP/designcrispr.html
CRISPR Protocol outline with Cas9 lentivirus:
1. select or design the 20bp target-sequence (crRNA) using an online CRISPR designer tool;
2. generate the gRNA lentivector and gRNA lentivirus: GenTarget provides gRNA lentivector cloning kits for you to construct your target specific gRNA lentivectors with the option to use H1 or U6 promoter, and the flexibility to use different selection markers.
GenTarget also provides services to generate your target-specific gRNA lentivirus, Please contact us for a service quote.
3. generate the Donor DNA by one of the methods listed below:
-
- method 1: synthesize the Double-stranded DNA cassette for sequence insertion:
” LHA (300bp loci specific left homologues arm) +(Promoter-insert-polyA terminator) + (RHA (300bp loci specific right homologues arm)”
-
- method 2: synthesize single-strand DNA oligo for knockOut:
” LHA (25 bp loci specific left homologues arm) +(target sequence with deletion or mutation) + (RHA (25bp loci specific right homologues arm)”
4. Add ready-to-use Cas9 expression lentivirus to target cells / organ;
5. Apply gRNA lentivirus,
6. Verify the target’s KnockOut;
7. For gene insertion or repair: deliver double-stranded Donor DNA into target cells / organ;
Gentarget Provides its proprietary technology for large gene repair and insertion, with high editing frequency.
8. Verification of the gene-editing results;
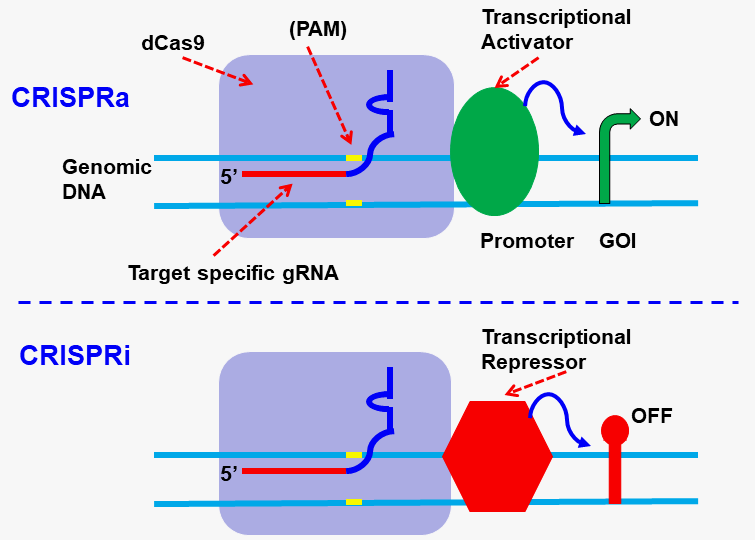
Epigenomic: CRISPRi and CRISPRa:
Gentarget Inc generated the dCas9 expression construct by making two point mutation in spCas9’s endonuclease domains (RuvC and HNH), as D10A and H840A, resulting in its deactivation.
The dCas9 protein binds to a specific genomic location under the guidance of a target site-specific gRNA. When coupled with a protein, the resulting “dCas9-Protein” fusion can modify the genome sequence by preventing the cell’s transcription machinery inducing CRISPR interference (CRISPRi) or activation (CRISPRa).
GenTarget’s CRISPR-Cas9 knock In/Out Services:
GenTarget can produce your desired reagents, for target-specific CRISPR/Cas9 genomic gene editing upon service requests. This service provides the most advanced reagents for carrying out your own genomic gene knock-In/Out experiments (or transgene animal models) in the chromosomal level. You do not need to buy or construct any lentivectors and no worry about each component’s delivery, which makes gene editing easier than ever. You simply add the provided reagents into your cells or organ.
To order it, you just select the target that you want to knock-Out and select the desired knock-In or repair gene or sequence, Gentarget will provide the following ready-to-use gene Knock-In/Out components:
- The nuclear penetrating hu Cas9 expression lentivirus with a desired antibiotic marker, or or Adenovirus, or Non-integrating Cas9 lentivirus;
- The target-specific “guide RNA lentivirus (gRNA)” lentivirus with the desired antibiotic selection marker, as well as a negative control gRNA lentivirus. We design the target-specific gRNA sequences, construct the gRNA expression lentivector under U6 or H1 promoter, produce the ready-to-use gRNA lentiviruses.
- Ready to use Donor cassette (the double-stranded DNA for knock-In or repair) with loci specific homologous arms: such as “RHA==insertion / repair ==LHA” ;
Please contact us for these service details.
References:
Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. (1987). “Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product“. Journal of bacteriology 169(12): 5429–5433. PMC 213968. PMID 3316184.
Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J. A.; Charpentier, E. (2012). “A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity”. Science 337 (6096): 816–821. doi:10.1126/science.1225829.PMID 22745249.